European Y-DNA and Mitochrondrial DNA (mtDNA) Haplogroups
Origins, Spread, and Ethnic Association of European Haplogroups and Subclades
Thia webpage from Eupedia.com – – Author – Maciamo Hay – adapted by Barry McKee.
Contents
- DNA Facts
- Introduction to Genetic Genealogy
- Haplogroups in Prehistoric Europe
- Y-DNA Haplogroups
- Chronological Development of Y-DNA Haplogroups
- Map of Early Bronze Age Cultures in Europe
- Y-DNA Haplogroups – with Focus on Europe and the Middle East
- Haplogroup A
- Haplogroup C
- Haplogroup E1b1
- Haplogroup G2a
- Haplogroup H
- Haplogroup I1
- Haplogroup I2
- Haplogroup J1
- Haplogroup J2
- Haplogroup L
- Haplogroup Mc
- Haplogroup Q
- Haplogroup R1a
- Haplogroup R1b – our McKee Haplogroup
- Haplogroup T
- Mitochondrial DNA (MtDNA) Haplogroups
- Chronological Development of mtDNA Haplogroups
- Chart of European mtDNA Haplogroups
- Mitochondrial DNA (mtDNA) of Prehistoric Europeans
- European mtDNA Haplogroups and their Subclades
- Haplogroups H and V (mtDNA)
- Haplogroups U and K (mtDNA)
- Haplogroups J and T (mtDNA)
- Haplogroup W (mtDNA)
- Haplogroup I (mtDNA)
- Haplogroup X (mtDNA)
- Haplogroup R (mtDNA)
- Finno-Uralic mtDNA
- Berber mtDNA
- Gypsy mtDNA
Disclaimer The information about the origin and ethnic association of haplogroups on this website should not be read as hard facts, but, as is often the case in science, as a model in constant evolution based on the present knowledge and understanding (of the author, Maciamo Hay). Whenever the advancement of genetics couldn’t provide irrefutable answers, we have attempted to provide the most likely and logical hypothesis based on archeological, historical and linguistic evidence.
DNA Facts
Nucleobases are the alphabet of DNA. There are four of them: adenine (A), thymine (T), guanine (G) and cytosine (C). They always go by pairs, A with T, and G with C. Such pairs are called “base pairs”.
The 46 chromosomes of human DNA are composed of a total of 3,000 million base pairs.
The Y chromosome possesses about 60 million nucleobases, and the X chromosome about 150 million bases.
Mitochondrial DNA (mtDNA) is inherited from mother to daughter. It is found outside the cell’s nucleus and is much smaller in size – about 16,000 bases.
Y-chromosomes contain sequences of repeating nucleotides known as short tandem repeats (STRs). The number of repetitions varies from one person to another. Individual Y-DNA sequences or STRs that have proven useful in genealogical DNA work are called markers, and each has a name. A SNP (single nucleotide polymorphism) is a mutation in a single base pair occurring commonly within a population. At present, only a few hundreds SNP’s define all the human haplogroups for mtDNA or Y-DNA.
Introduction to Genetic Genealogy
DNA studies have permitted us to categorise all humans on Earth in genealogical groups sharing one common ancestor at one given point in prehistory. They are called Haplogroups , and their subgroups are Subclades. There are two kinds of haplogroups: the paternally inherited Y-chromosome DNA (Y-DNA) haplogroups, and the maternally inherited mitochondrial DNA (mtDNA) haplogroups. They respectively indicate the agnatic (or patrilineal) and cognatic (or matrilineal) ancestry.
Y-DNA haplogroups are useful to determine whether two apparently unrelated individuals sharing the same surname do indeed descend from a common ancestor in a not too distant past (3 to 20 generations). This is achieved by comparing the haplotypes through the STR markers.
Deep SNP testing allows to go back much farther in time, and to identify the ancient ethnic group to which one’s ancestors belonged (e.g. Celtic, Germanic, Slavic, Greco-Roman, Basque, Iberian, Phoenician, Jewish, etc.).
In Europe, mtDNA haplogroups are quite evenly spread over the continent, and therefore cannot be associated easily with ancient ethnicities. However, they can sometimes reveal some potential medical conditions. Some mtDNA subclades are associated with Jewish ancestry, notably K1a1b1a, K1a9,d K2a2a and N1b.
The study of Y-chromosomes is more interesting than that of mitochondrial DNA for two reasons.
Firstly, the Y chromosome is a sequence of 60 million “characters” (nucleobases), against only 16,569 for mtDNA. The Y chromosome therefore offers a much greater resolution as mutations are more common, and indeed happen pretty much every generation. In contrast, mtDNA mutations happen much more infrequently. Since the time of the Mitochondrial Eve, approximately 200,000 years ago, modern humans have acquired in average 20 mtDNA mutations in each lineage – about one every ten thousand years. Even though the number of mutations has accelerated with the soaring of human population over the last 10,000 years, the dating of lineages based on mtDNA alone remains very approximate, and practically useless for historical times./
By sequencing the full Y chromosome, it is theoretically possible to map the entire patrilineal genealogy of humanity (or any other species) within a few generations (in some cases even within one generation). This is a colossal task, and expensive one too, since full chromosome sequencing (reading every nucleobase one by one) remains very expensive compared to SNP genotyping (checking only for mutations already discovered in other individuals). DNA tests provided to the general public (23andMe, FTDNA…) only use genotyping, so new mutations are not normally discovered by such tests (unless they are repeat mutations from other haplogroups). This is why population geneticists have only managed to sketch very broad lineages so far. The deepest subclades identified still encompass tens or hundreds of thousands of individuals.
The second advantage of Y-DNA over mtDNA is that men have traditionally been less mobile than women. In almost every settled, agricultural society, men are the ones who inherit their parents’ property, and therefore remain in the same location generation after generation. Women, on the other hand, were often send away to marry in another village or town, so that their lineages spread more evenly over time. < ?p>
Paternal and Maternal Haplogroups in Prehistoric Europe
Mesolithic Europe
See the Timeline of Human Prehistory for information on the Mesolithic period. Following the end of the last Ice Age approximately 12,000 years ago, European hunter-gatherers recolonized the continent from the Ice Age refugia in southern Europe. The vast majority of Mesolithic Europeans would have belonged to Y-haplogroup I. This included I*, pre-I1, I1, I2*, I2a*, I2a2, but the most widespread appears to have been I2a1, which was found in most parts of Europe. Northeast Europeans would have belonged mostly to haplogroup R1a. Other minor male lineages were certainly also present in parts of Europe, notably haplogroup A1a, C-V20, F-P96 and possibly even Q1a and R1b1* (P25). The maternal lineages of Mesolithic Europeans appears to have been predominantly U4 and U5, but also included several H subclades (H1, H3, H17), T, U2 and V. The presence of mt-haplogroups I and W is likely but hasn’t been confirmed yet.
Based on their modern distributions, mtDNA haplogroups H10 and H11 might well have Mesolithic/Palaeolithic European origins. There seem to have been several Palaeolithic and/or Mesolithic migrations from Northwest Africa to Iberia. The oldest might have brought West African haplogroup A1a to Western and Northern Europe during the Palaeolithic. A1a has been found in modern populations as far north as Ireland, Scotland, Scandinavia and Finland. E-M81 probably crossed the Gibraltar Straits some time during the Mesolithic and settled around most of Iberia, probably expanding into the rest of Western Europe during the Neolithic period (Megalithic and Bell Beaker cultures). The presence of African maternal lineages (L2, L3 and possibly L1b1) has been attested in Neolithic Iberia. Northwest Africans would also have brought U6 and possibly HV0/V lineages to Europe.
A small percentage of sub-Saharan African admixture has been identified in Late Mesolithic Swedes from the Pitted Ware culture (2800-2000 BCE), which would imply that A1a was already present in northern Europe at the time. Another Mesolithic sample from Loschbour in Luxembourg had dark hair and considerably darker skin than modern Europeans.
Neolithic and Chalcolithic Europe
See the Timeline of Human Prehistory for information on the Neolithic period; the Chalcolithic (Copper Age) followed. Agriculture first developed in the Levant, then spread to Anatolia, Greece, the Balkans, Italy, Central and Eastern Europe. These Neolithic farmers were confirmed to have belonged to Y-DNA haplogroups E1b1b and G2a, but probably also included a minority of J1, J2 and T lineages.
Hundreds of Neolithic samples from all over Europe (but especially Central Europe and Iberia) have been tested. The new lineages brought by these Near Eastern immigrants included mt-haplogroups HV, J1, J2, K1, K2, N*, N1, T1a, T2b, T2c, T2e, T2f, U3, W, X1, X2, and many subclades of H (including H2, H5, H7, H13 and H20). H4, H8 and H9 seem to have originated in the Near East as well, although no Neolithic sample has been identified in Europe yet.
However, due to the proximity of the Caucasus from the Indo-European homeland, many of these mt-haplogroups were almost certainly also transported by the Indo-Europeans themselves, notably H5, K1a, T2b, U3, W and X2.
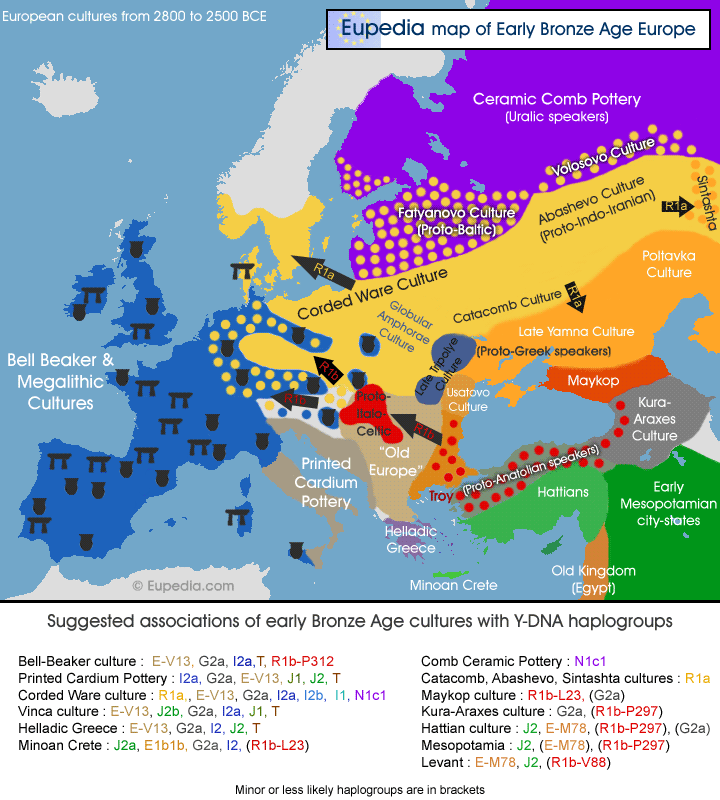
The Bronze Age and the Indo-European migrations
The origin of the Indo-Europeans lies in the Pontic-Caspian steppe with (R1a) tribes to the north (forest-steppe and tundra) and (R1b) tribes to the south (open steppe) during the Chalcolithic and Bronze Age. Their migration both westward to Europe and south-eastward to Central and South Asia makes it easy to guess which mtDNA haplogroups they carried The best matches for R1a are C4a, H1b, H1c, H2a1, H6, H11, K1b1b, K1c, K2b, T1a1a1, T2a1b1, T2b2, T2b4, U2e, U4, U5a1a, W, and several I subclades.
The R1b branch would have originated in eastern Anatolia and/or northern Mesopotamia/Syria during the Early Neolithic period, where they probably domesticated cattle and became primarily cattle herders. Then would have crossed the Caucasus to the Pontic Steppe in search for pasture for their cattle, where they mixed to some extent with southern R1a tribes. The maternal lineages of these Near Eastern R1b people would have included haplogroups H5a, H6, H8, H15, I1a1, J1b1a, K1a3, K2a6, U5, and some V subclades (like V15).
mtDNA haplogroups H4 has not been found in Europe before the Late Chalcolithic (Corded Ware) and Bronze Age (Unetice) and might have been brought by the Indo-Europeans. Likewise, H6 is absent from all Mesolithic or Neolithic samples, and its strong presence in the North Caucasus and Central Asia supports an Indo-European connection. Haplogroup V has never been found in prehistoric sites in Northeast Europe, nor in any Indo-European burial in the Eurasian steppe or Central Asia. It is nevertheless present in every part of Europe nowadays. Its frequency is higher than the European average in north-western Russia (> 5%), and peaks among the Sami (> 30%).
Haplogroup V has also been found in most Uralic and Altaic populations across North Asia, and at trace frequencies as far as Korea and Japan. More intriguingly, haplogroup V is one of the four Eurasian haplogroups found among the Fulani people of Central Africa, who have high percentages of haplogroup R1b-V88. It is therefore likely that V was one of the original haplogroups of R1b people, and perhaps of the Paleolithic mammoth hunters from whom R1b is descended. Some V lineages could have been absorbed by the expansion of Ural-Altaic populations (Y-haplogroup N) in North Asia, which would explain its high frequency among the Finns and Sami.
Y – DNA Haplogroups
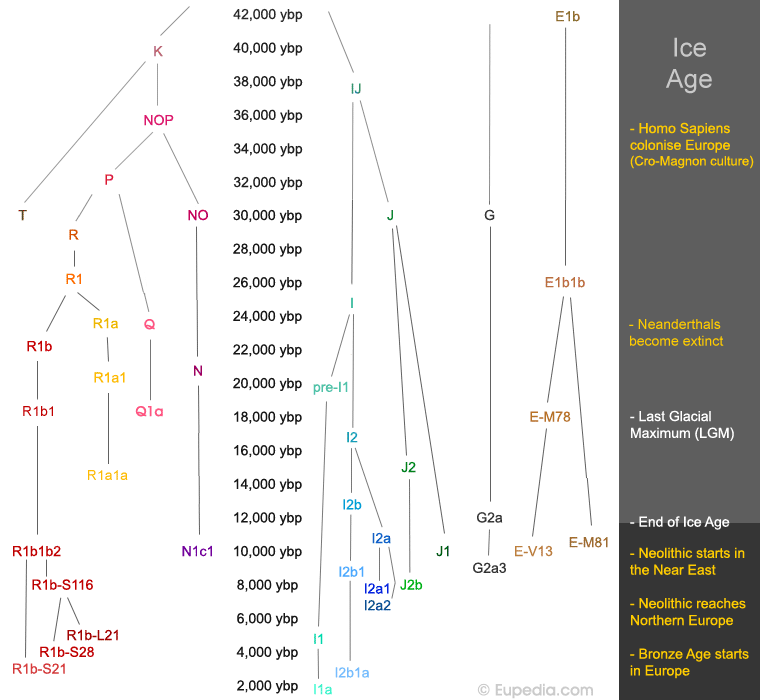
Figure: Chronological Development of Y- DNA Haplogroups
K => 40,000 years ago (probably arose in northern Iran)
T => 30,000 years ago (around the Red Sea or around the Persian Gulf)
J => 30,000 years ago (in the Middle East)
R => 28,000 years ago (in the Central Asia)
E1b1b => 26,000 years ago (in Northeast Africa)
I => 25,000 years ago (in the Balkans)
J1 => 20,000 years ago (in the Taurus/Zagros mountains)
J2 => 19,000 years ago (in northern Mesopotamia)
E-M78 => 18,000 years ago (in north-eastern Africa)
R1b => 18,000 years ago (around the Caspian Sea or Central Asia)
R1a => 17,000 years ago (in southern Russia)
G => 17,000 years ago (in the Middle East)
I2 => 17,000 years ago (in the Balkans)
E-V13 => 15,000 years ago (in the southern Levant or North Africa)
I2b => 13,000 years ago (in Central Europe)
N1c1 => 12,000 years ago (in Siberia)
E-M81 => 11,000 years ago (in Northwest Africa)
I2a => 11,000 years ago (in the Balkans)
G2a => 11,000 years ago (in the Levant or Anatolia)
R1b1b2 => 10,000 years ago (north or south of the Caucasus)
I2b1 => 9,000 years ago (in Germany)
I2a1 => 8,000 years ago (in Southwest Europe)
I2a2 => 7,500 years ago (in Southeast Europe)
I1 => 5,000 years ago (in Scandinavia)
R1b-L21 => 4,000 years ago (in Central or Eastern Europe)
R1b-S28 => 3,500 years ago (around the Alps)
R1b-S21 => 3,000 years ago (in Frisia or Central Europe)
I2b1a => less than 3,000 years ago (in Britain)
Map of Early Bronze Age Cultures in Europe around 4,500 to 5,000 years ago
Y-DNA Haplogroups – with Focus on Europe and the Middle East
Haplogroup A (Y-DNA)
A is the oldest of all Y-DNA haplogroups. It originated in sub-Saharan Africa over 140,000 years ago, and possibly as much as 340,000 years ago if we include haplogroup A00. Modern populations with the highest percentages of haplogroup A are the Khoisan (such as the Bushmen) and the southern Sudanese.
There are only rare cases of European men belonging to haplogroup A – all belonging to the same A1b1b2 (M13) subclade, which is normally found in East Africa (Ethiopia, Sudan), but has also been found in Egypt, the Arabian peninsula, Palestine, Jordan, Turkey, Sicily, Sardinia and Algeria. It was brought to Europe by Levantine people, during the Neolithic or later times(Phoenicians, Jews, immigration within the Roman Empire).
Haplogroup A1a* (M31) has been found in Finland, Norway and eastern England. This subclade is normally found along the west coast of Africa (Guinea-Bissau, Cape Verde, Mali, Morocco) and could have come to Europe during the Paleolithic.
Haplogroup C (Y-DNA)
Haplogroup C is an extremely old lineage thought to have appeared before or soon after the first migration of Homo Sapiens outside Africa, some 70,000 years ago. Men belonging to haplogroup C would have departed from East Africa during the Ice Age and followed the coasts of Indian Ocean, settling in the Arabian peninsula, the Indian subcontinent, south-east Asia, north-east Asia and Oceania. The first group to split away was C-Z1426, which colonised the Middle East and South Asia. One branch (CTS11043) might have moved north to Central Asia, then split into two: one tribe moving west to Europe (haplogroup C-V20) while the other migrated to East Asia and survives only in Japan today (haplogroup C-M8). Haplogroup C-V20 probably represents the first migration of Homo Sapiens to Europe 45,000 years ago, and would therefore have been the first to come into contact with Neanderthals.
The second branch of C-Z1426 spread around South Asia, Southwest Asia, and Central Asia, where it is found at low frequencies nowadays (haplogroup C-M356).
During that time, other C tribes continued their eastward migration to south-east Asia, where they split in four main regional clusters. The first branch colonised Indonesia, Melanesia, Micronesia, and Polynesia (haplogroup C2-M38). A second branch would have gone south to Australia, where they became the Aborigines (haplogroup C4-M347). Another settled in the highlands of New Guinea (haplogroup C-P55). The fourth branch went all the way up the north-east Asia (haplogroup C3-M217) and is found nowadays chiefly among the Mongols, tribes descended from the Mongols (Kalmyks, Hazaras) including Turkic people (Kazakhs, Kyrgyz, Uyghurs, Uzbeks, Tuvans, Yakuts), East Siberian tribes (Buryats, Chukchi, Itelmens, Nivkh, Tungusic peoples), Chinese (Han, Hui, Manchus, Oroqens, Tujia), Koreans and Japanese (especially the Ainus), but also among several indigenous peoples of North America, including some Na-Dené-, Algonquian-, or Siouan-speaking populations.
Haplogroup C is a very rare lineage in Europe. The few Europeans who belong C either belong to the European C-V20, the Middle Eastern C-M358, or the Mongolian C3-M217. Haplogroup C3 has also been identified in one Hunnic skeleton from the Iron Age in present-day Mongolia. Its presence in Europe can therefore be linked to the Hunnic and Mongolian invasions, like haplogroup Q1a.
Haplogroup E1b1b (Y-DNA)
The main paternal lineage of North Africa. It is linked to the diffusion of Afro-Asiatic languages and of Neolithic farmers from the Near East and the Balkans.
Haplogroup G2a (Y-DNA)
An important paternal lineage of Neolithic farmers and herders in the Middle East and Europe. It is also associated with the Indo-Europeans.
Haplogroup H (Y-DNA)
Haplogroup H is typically found among Dravidian populations in the Indian subcontinent, especially in South India and Sri Lanka. In Europe it is found almost exclusively among the Gypsies (Romani), who belong predominantly (between 15% and 50%) to the H1a (M82) subclade of Indian origin. The highest frequencies of haplogroup H among non-Romani Europeans are found in regions with large Romani populations, such as Romania, Slovakia, the southern Balkans, and Andalusia, suggesting that these lineages are also of Romani origin. No other subclade than H1a has been found to date in Europe.
Haplogroup I1 (Y-DNA)
The original paternal lineage of Nordic Europe. It was propagated around Europe by ancient Germanic tribes and the Vikings.
Haplogroup I2 (Y-DNA)
Continental Europe’s largest Mesolithic paternal lineage. It is common among Slavic and Germanic people.
Haplogroup J1 (Y-DNA)
The dominant Arabic and Cohanim paternal lineage. It originated around the Caucasus and expanded during the Neolithic towards East Africa.
Haplogroup J2 (Y-DNA)
A major West Asian and Greco-Roman paternal lineage. It is associated with many Bronze and Iron Age ancient civilizations from the Mediterranean to India.
Haplogroup L (Y-DNA)
Geographic distribution –
Haplogroup L is found mostly in West Asia and South Asia. Its overall frequency ranges between 5 and 15% in Pakistan and western India, and from 1 to 10% in central Asia (mostly in Uzbekistan, Tajikistan and Afghanistan). It is also found in the Middle East (5% in Lebanon, 4.5% in Turkish Kurdistan, 4% in Iran, 3% in Syria), in parts of the Caucasus (7% in Azerbaijan and Chechnya, 3% in Armenia and Ingushetia), and in isolated parts of Europe (3.5% in north-east Italy, from 0.2% to 1% in the Balkans and Greece, 0.5% in Flanders).
Subclades –
Haplogroup L is divided in four main subclades:
L1a (M27) is the mostly found in India and Sri Lanka, with frequencies decreasing towards Pakistan, southern Iran, the Arabian peninsula. It has also been found in Piedmont (Italy), Rhineland (Germany) and Flanders (Belgium) .
L1b (M317) is found chiefly in the South Caucasus, eastern Anatolia and Lebanon. It has also been found in South Tyrol, Russia and Central Asia. Its main subclade L1b1 (M349) has been found in Italy, Switzerland, Austria, Germany, Belgium, England, northern Ireland, and scattered around most of central and eastern Europe and the eastern Mediterranean. The presence of L1b and L1b1 in Europe probably dates back to the Neolithic period.
L1c (M357) is an essentially Gedrosian subclade, found among the Burushos, Kalashs and Pashtuns of Pakistan and Afghanistan, but also among the Chechens in the north-east Caucasus. It is also found at low frequencies in other populations of Pakistan, in India, northern Iran, Georgia and Ingushetia. In Europe it has been found in Sicily.
L2 (L595) has been found exclusively in Europe (Greece, Italy, southern Germany, Russia) and in the South Caucasus.
Haplogroup N1c (Y-DNA)
The main Uralic lineage, found chiefly among the Finns, Sami, Volga-Ural ethnic groups and Baltic people, and to a lower extent also Slavic and Germanic people.
Haplogroup Q (Y-DNA)
A North and Central Asian lineage linked to the Huns, Mongols and Turkic people. It is also a minor Scandinavian and Jewish lineage.
Haplogroup R1a (Y-DNA)
The dominant paternal lineage in Northeast Europe and southern Central Asia. It was diffused around Eurasia by the Indo-Aryans and Balto-Slavic people.
Haplogroup R1b (Y-DNA) – our McKee Haplogroup
The dominant paternal lineage in Western Europe. It represents the Greco-Anatolian, Italic, Celtic and Germanic branches of the Indo-European speakers. We now know that our McKee Family belongs to this R1b Haplogroup, and we have a subpage with R1b History.
Haplogroup T (Y-DNA)
An old Middle Eastern and East African lineage. It was found among ancient Babylonians, Phoenicians, Greeks and Romans.
Mitochondrial DNA (MtDNA) Haplogroups
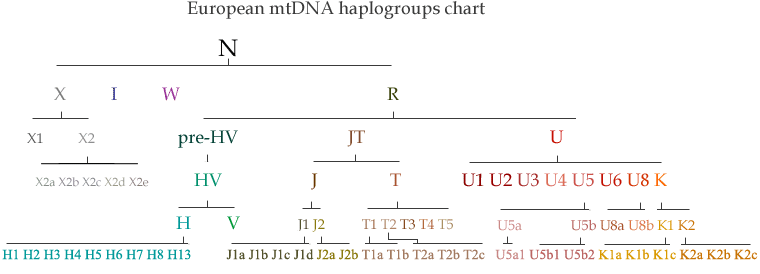
All mtDNA haplogroups found in Europe descend from the N group, which is thought to represent one of the two initial migrations by modern humans out of Africa, some 60,000 to 80,000 years ago. Nowadays haplogroup N is only found at extremely low frequencies in various parts of Eurasia.
Unfortunately, the tiny size of mitochondrial DNA (approximately 16,500 base pairs as opposed to 60 million for Y-DNA) does not allow a very accurate tracing of ancestry. Basal mitochondrial haplogroups all arose during the Ice Age, a period when humans were nomadic hunter-gatherers, well before the establishment of cities and civilizations. Even deep subclades generally point to a common Neolithic or Bronze Age ancestry, but rarely later than that, and do not necessarily match any recognisable historical ethnic and linguistic groups. One likely reason is that women, through whom mtDNA is passed, tended to marry outside their ethnic group more often than men (e.g. to secure an alliance between two tribes or kingdoms). Haplogroups associated with European or Middle Eastern descent are H, I, J, K, T, U, V, W and X (except the branch X2a which found among Native Americans).
Chronological Development of mtDNA Haplogroups
Note that the age of mitochondrial haplogroups is much more difficult to estimate than Y-DNA haplogroups, due to the tiny sequence of mtDNA and the few number of mutations available. The error margin for the dates below is typically of +/- 5,000 years, but could even exceed that for older haplogroups.
N => 75,000 years ago (arose in North-East Africa)
R => 70,000 years ago (in South-West Asia)
U => 60,000 years ago (in North-East Africa or South-West Asia)
pre-JT => 55,000 years ago (in the Middle East)
JT => 50,000 years ago (in the Middle East)
U5 => 50,000 years ago (in Western Asia)
U6 => 50,000 years ago (in North Africa)
U8 => 50,000 years ago (in Western Asia)
pre-HV => 50,000 years ago (in the Near East)
J => 45,000 years ago (in the Near East or Caucasus)
HV => 40,000 years ago (in the Near East)
H => over 35,000 years ago (in the Near East or Southern Europe)
X => over 30,000 years ago (in north-east Europe)
U5a1 => 30,000 years ago (in Europe)
I => 30,000 years ago (Caucasus or north-east Europe)
J1a => 27,000 years ago (in the Near East)
W => 25,000 years ago (in north-east Europe or north-west Asia)
U4 => 25,000 years ago (in Central Asia)
J1b => 23,000 years ago (in the Near East)
T => 17,000 years ago (in Mesopotamia)
K => 16,000 years ago (in the Near East)
V => 15,000 years ago (arose in Iberia and moved to Scandinavia)
H1b => 13,000 years ago (in Europe)
K1 => 12,000 years ago (in the Near East)
H3 => 10,000 years ago (in Western Europe)
Chart of European mtDNA Haplogroups
Mitochondrial DNA (mtDNA) of Prehistoric Europeans
The testing of ancient mtDNA help us to understand how long each haplogroup has been in Europe. Dozens of samples from the Paleolithic and Mesolithic, and hundreds from the Neolithic, Chalcolithic and Bronze Age have already been tested. Eupedia.com provides a list,Prehistoric European mtDNA by period and culture.
European mtDNA Haplogroups and their Subclades
Haplogroups H and V (mtDNA)
We see in the chart above that H and V are subclades of HV. Haplogroup H is by far the most common all over Europe, amounting to about 40% of the European population. It is also found (though in lower frequencies) in North Africa, the Middle East, Central Asia, Northern Asia, as well as along the East coast of Africa as far as Madagascar.
H1, H3 and V are the most common subclades of HV in Western Europe. H1 peaks in Norway (30% of the population) and Iberia (18 to 25%), and is also high among the Sardinians, Finns and Estonians (16%), as well as Western and Central European in general (10 to 12%) and North-West Africans (10 to 20%). H3 is commonest in Portugal (12%), Sardinia (11%), Galicia (10%), the Basque country (10%), Ireland (6%), Norway (6%), Hungary (6%) and southwestern France (5%). Haplogroup V reaches its highest frequency in northern Scandinavia (40% of the Sami), northern Spain, the Netherlands (8%), Sardinia, the Croatian islands and the Maghreb. It is likely that H1, H3 and V, along with haplogroup U5, were the main haplogroups of Western European hunter-gatherers living in the Franco-Cantabrian refuge during the last Ice Age, and repopulated much of Central and Northern Europe from 15,000 years ago. Haplogroup H13 is most common in Sardinia and around the Caucasus. Its distribution is reminiscent of Y-DNA haplogroup G2a. The same is true of H2 to a lower extent. This would suggest a Caucasian or Anatolian origin.
H5 and H7 are also common in the Caucasus, but their lower incidence around the Mediterranean, and higher frequency from Anatolia to the Alps via the Danube suggest a possible link with the spread of agriculture (YDNA E1b1b, J2 and T) or of the Indo-Europeans (R1b1b2).
Haplogroups U and K (mtDNA)
Haplogroup U is extremely old. It originated some 60,000 years ago at the confine of North-East Africa and the Middle East, soon after the first Homo Sapiens ventured out of Africa. This is why each of its top-level subclade (U1, U2, U3…) can be seen as a haplogroup in its own right. The main European subclades are U3, U4, U5 and U8/K. U1 is mostly found in the Middle East, U6 in North Africa, U7 from the Near East to India, and the rare U9 from Ethiopia and the Arabian peninsula to Pakistan.
Haplogroup U2 is found primarily in South Asia, but probably is of Indo-European origin as it is found at low frequencies throughout the Pontic-Caspian steppe and has been identified in a 30,000 year-old Cro-Magnon from the middle Don valley in Russia. It might have been the dominant haplogroup of the northern forest-steppe foragers who later became the Proto-Indo-Iranian speakers (see R1a above) and moved massively to Central and South Asia.
Haplogroup U3 is centered around the Black Sea, with a particularly strong concentration in the north-eastern part. It could be related to the ancient Indo-Europeans, and probably more to R1b than R1a.
Haplogroup U4 is strongly associated with Y-haplogroup R1a. It is found in most of Europe, but especially in Balto-Slavic countries, but also in Siberia, Central Asia, Afghanistan and northern Pakistan. U4 was already present in many parts of Europe (Russia, Sweden, Germany, Portugal) during the Mesolithic period, but seems to have almost disappeared from central Europe during the Neolithic, before being re-introduced by the Proto-Indo-European speakers from Russia and Ukraine during the Bronze Age.
Haplogroup U5 is the most common in Western and Northern Europe. DNA tests on ancient skeletons have shown that U5 was the principal mitochondrial haplogroup of Paleolithic and Mesolithic hunter-gatherers in Northern Europe. Ancient DNA tests conducted in Britain, Germany and Scandinavia indicate that the frequency of U5 has progressively declined over time through the Neolithic, Bronze Age, Iron Age and Middle Ages. Nowadays it remains most common in the far north of Europe, where the Mesolithic population has been least affected by subsequent migrations. For instance, 30 to 50% of the Sami people of northern Scandinavia belong to haplogroup U5b (and about 40% to haplogroup V, which is also pre-Neolithic European origin). Haplogroup K is the main subclade of U8. It is found throughout Europe and Western Asia, as far away as India. Its highest concentration is in North-West and Central Europe, Anatolia and the southern Arabian peninsula. It is believed to have first arisen somewhere between Egypt and Anatolia approximately 16,000 years ago (estimates range from 22,000 years to as little as 10,000 years before present). It has the largest number of subclades of any haplogroup in spite of its fairly recent age. K1a is the largest subclade. The relatively important presence of K1a in the Near East suggest that it predates the Neolithic migration to Europe. This has been supported by the ancient mtDNA from Neolithic sites. Haplogroup K was never found in Europe prior to the Neolithic, then suddenly appears at a frequency (17%) much higher than in modern Europeans and similar to that of the present-day Levant. Most of the Neolithic K belongs to the K1a subclade.
Most K1a4, K1a10, K1b, K1c and K2 subclades are typically European. K1a4 is also common in Anatolia and Greece, and could indeed have spread to the rest of Europe from there during the Neolithic period, along with haplogroups J and T (and Y-DNA haplogroups E1b1b, J2 and T). The Indo-Europeans from Anatolia could also have contributed to the propagation of K. K1a1b1a and K1a9 are found primarily among Ashkenazi Jews.
Haplogroups J and T (mtDNA)
Haplogroup J originated in the Middle East 45,000 years, making it one of the oldest mitochondrial haplogroups in Europe and the Middle East. Haplogroups J1c and J2a1 might have been present in Southeast Europe since the Epipaleolithic, then were probably diffused by Neolithic farmers across the rest of Europe. J2b1a, a mostly Near Eastern subclade, has been found in Neolithic samples in Europe alongside J1c.
Haplogroup J1b is found across the Near East, particularly between the Caucasus, Iran and Arabia. J1b1a is the only J1b subclade typically found among Europeans. It is present all over Europe as well as around the Caucasus, in Central Asia and the Altai, and was almost certainly spread by the R1b branch of the Indo-Europeans. J1d, J2a2, J2b2 are essentially confined to the Middle East (+ North Africa for J2a2).
Haplogroup T is thought to have originated in the Middle East about 30,000 years ago. It is found throughout Europe, the northern half of Africa through the Near East to Central Asia and Siberia, with pockets in India and North-West China (Xinjiang). Some T1 and T2 subclades are thought to have entered Europe during Late Glacial and the immediate postglacial periods, but to have been dispersed around Europe mostly by later population movements, first with agriculturalists during the Neolithic, then with the Proto-Indo-European speakers from the Pontic Steppe during the Bronze Age.
Haplogroups W (mtDNA)
Present at low frequencies in most of Europe, in Anatolia, around the Caspian Sea, and from the Indo-Pakistani border to Xinjiang, haplogroup W is one of the best maternal markers of Indo-European ancestry (mtDNA equivalent of R1a and R1b). Its highest frequency is in Ukraine, European Russia, Baltic countries and Finland (3 to 5% overall), as well as in northern Pakistan (15%), Punjab (9%) and Gujarat (12%). In India, it is considerably more common among the upper castes and among Indo-European speakers.
Haplogroups I (mtDNA)
Like haplogroup W, haplogroup I is found at low frequency over most of Europe, especially in northern and eastern Europe, and across Central Asia as far as Pakistan and North-West India, with a characteristic presence in the North Caucasus. Haplogroup I first appears in Europe with the arrival of Proto-Indo-European cultures, notably the Unetice culture associated with Y-haplogroup R1b. The absence of haplogroup I from Paleolithic, Mesolithic and Neolithic sites, and from modern non-Indo-European speaking populations such as the Saami, the Basques and the Maghrebians all play in favour of an Indo-European origin.
Haplogroups X (mtDNA)
Haplogroup X is a very old and scattered haplogroup found all over Eurasia, North Africa as well as among Native North Americans. It frequency rarely exceeds 5% of the population in any ethnic group, and is more often restricted to 1 or 2%. X1 is found almost exclusively in North Africa, while X2a is the only lineage present among Amerindians. X2d, X2e, X2n and X4 are found in Europe and Central Asia, and could therefore have been spread at least partially by the Proto-Indo-Europeans. The strong presence of X2 around the Caucasus, progressively fading towards the Near East and Mediterranean, hints that it could be related to the spread of Y-DNA haplogroup G2a. R1b1b and G2a both having origins around the Caucasus it is unsurprising to find X2 alongside these two Y-DNA haplogroups.
Haplogroups R (mtDNA)
Haplogroup R is the main subclade of N, the one that was to generate the 6 most common European haplogroups (H, V, J, T, U, K). At the time of writing R subclades were numbered from R0 (a.k.a. pre-HV) to R31. Most of them are found in South Asia (R5, R6, R7, R8, R30, R31), Southeast Asia (R9, R21, R22, R24), East Asia (R9/F, R11/B), and even among Papuans (R14) and Australian aborigines (R12). R0a peaks in the southern Arabian peninsula is common among Arabs and Middle-Easterners. R1a (not to be confused with the homonymous Y-chromosome haplogroup) is found among the Adygea people from the North Caucasus (related to the Maykop culture), Brahmins from northern India, northwestern Russians and Poles – basically all people closely related with the Indo-European expansion. R2 is found from northwest India and Pakistan to Iran, Georgia and Turkey. It could be connected to the Indo-Iranians.
Finno-Uralic mtDNA
Finno-Uralic people have an overall mtDNA admixture similar to other Europeans, with a higher percentage of W and U5b, and a small percentage of Siberian haplogroups such as N or A. The Sami are characterised by a high percentage of haplogroups U5b1 and V.
Berber mtDNA
The Berbers are the indigenous population of north-west Africa. Although their Y-DNA is almost perfectly homogenous, belonging to haplogroup E-M81, Berber maternal lineages show a much greater diversity, as well as regional disparity. At least half (and up to 90% in some regions) of the Berbers belong to some Eurasian lineages, such as H, HV, R0, J, T, U, K, N1, N2, and X2, mostly of Middle or Near Eastern origin. 5 to 45% of the Berbers will have sub-Saharan mtDNA (L0, L1, L2, L3, L4, L5). There are only three native North African lineages, U6, X1 and M1, representing 0 to 35% of the people depending on the region.
Haplogroup U6 has been observed from the Iberia and the Canary Islands to Senegal in the West, and from Syria to Ethiopia and Kenya in the East. It is also found at low density in Europe, though mostly limited to Iberia. Approximately 10% of all North Africans belong to this lineage.
Gypsy mtDNA
The Gypsies (Romani people) originated in the Indian subcontinent and mixed with local population in the Middle East and Eastern Europe over the centuries. About half of the Gypsy population belong to haplogroup M, and more specifically M5 (reflected by Y-haplogroup H1a), which is otherwise exclusive to South Asia. The other mtDNA haplogroups found among the Gypsy community are mostly of Eastern European, Caucasian or Middle Eastern origin, such as H (H1, H2, H5, H9, H11, H20, among others), J (J1b, J1d, J2b), T, U3, U5b, I, W et X (X1b1, X2a1, X2f). The same diversity exist on the Y-DNA side (45% of H1a, followed by I1, I2a, J2a4b, E1b1b, R1b1b, R1a1a).
Books on European Prehistory
The Horse, The Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World, by David Anthony
The Lost World of Old Europe, by David Anthony
Encyclopedia of Prehistory: Europe, Volume 4, By Peter Neal Peregrine and Melvin Ember
Useful links
ISOGG (International Society of Genetic Genealogy)
The Journey of Mankind : interactive map of the spread of humans on Earth
Y-search : public Y-DNA database
Mitosearch : public mtDNA database
SNPedia : human DNA mutations database
Y-full : latest Y-DNA tree with mutations
PhyloTree : complete mtDNA tree with mutations
Y-DNA haplogroup predictor (based on STR markers)
Eupedia.com